ENCD(http://www.bio-server.cn/ENCD/) is a manually curated database that provides comprehensive experimentally supported associations about endocrine system diseases (ESDs) and long non-coding RNAs (lncRNAs). The incidence of ESDs has increased in recent years often accompanying other chronic diseases and can lead to disability. A growing body of research suggests that lncRNA plays an important role in the progression and metastasis of ESDs. However, there are no resources focused on collecting and integrating the latest and experimentally supported lncRNA-ESD associations. Hence, we developed an ENCD database that consists of 1379 associations between 35 ESDs and 501 lncRNAs in 12 human tissues curated from 353 literature. By using ENCD, users can explore the genetic data for diseases corresponding to body parts of interest as well as study the number of entries for ESDs of interest and all the corresponding disease data. ENCD also provides a flexible tool to visualize a disease-centered or gene-centered regulatory network. In addition, ENCD offers a submission page for researchers to submit their newly discovered endocrine disorders-genetic data entries online. Collectively, ENCD will provide comprehensive insights for investigating the ESDs associated with lncRNAs.
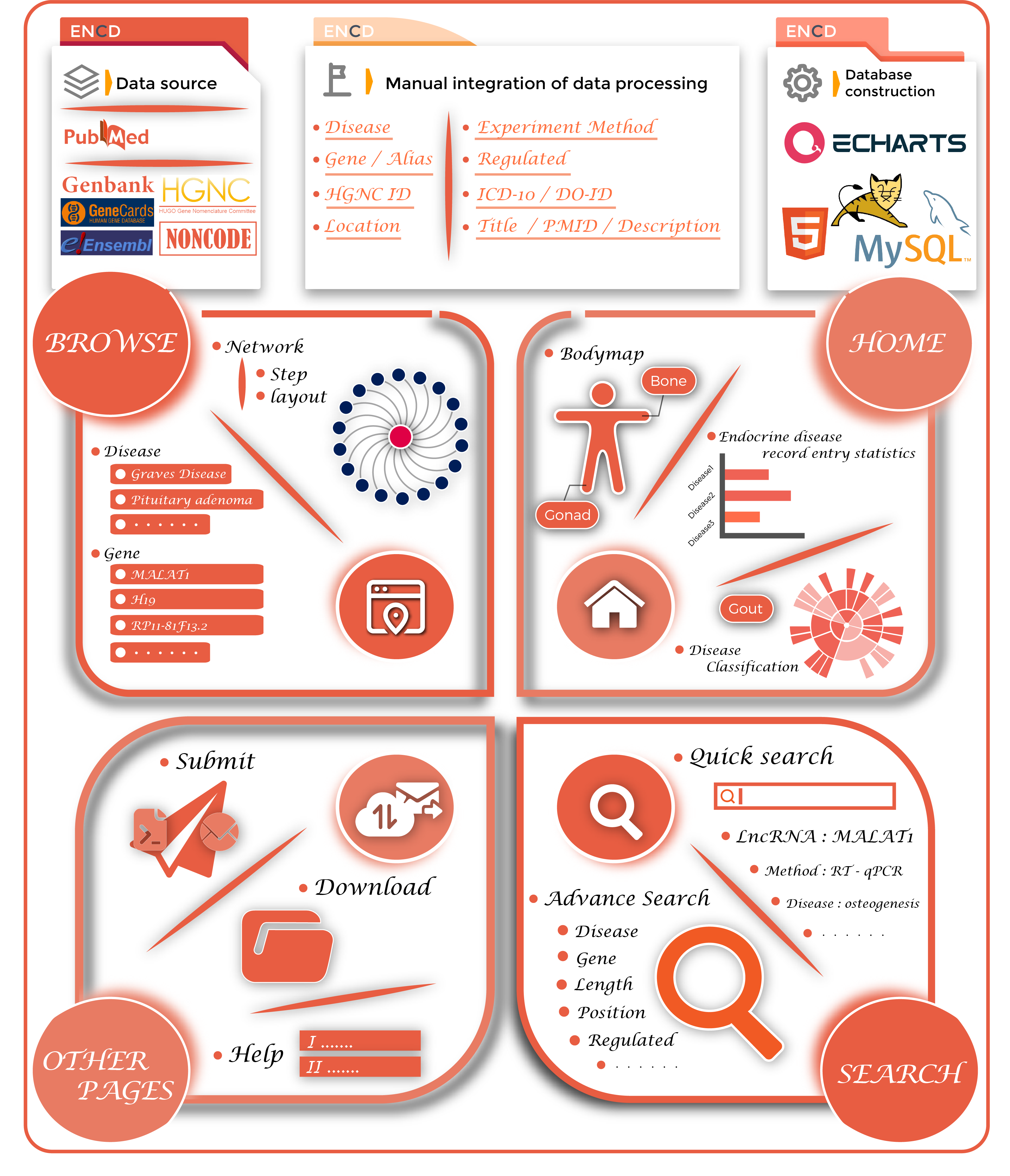
ENCD aims to collect and integrate information on all lncRNAs associated with endocrine diseases and its data sources are broadly divided into two categories. One category is derived from other databases, such as Noncode(1), Gencards(2), Ensembl(3), HGNC (4), Genbank(5), etc. The other category was derived from work already published by previous studies, and all literature related to endocrine disorders in recent years was selected by searching the PubMed database(6) with a list of keywords, such as ‘long non-coding RNA’, ‘Diabetes’, ‘pituitary dwarf’ and ‘multiple endocrine neoplasia type 1’.We collected detailed information of lncRNA-disease associations, including official names, synonyms, gene IDs, sequence and positional information of the lncRNAs, experimental techniques (e.g., microarray, qRT-PCR), regulating directions (up- or down-expression of lncRNAs), literature information (PubMed ID, year of publication and title of paper) and a brief functional description of lncRNA-disease associations from the original studies. According to relationships between experimental types and data, the type of disease-lncRNA associations were grouped into four categories, including Strong evidence, Weak evidence, Direct evidence, and High-throughput. This strategy has been used to mine miRNA-target (7) and cancer-lncRNA (8) associations. Strong evidence refers to the evidence of the correlation between LncRNA and diseases obtained through qRT-PCR, RNA interference, in vitro knockdown, and other laboratory technologies. Weak evidence refers to the prediction of disease candidate lncRNA through microarrayand machine learning methods. Direct evidence refers to a direct interaction between lncRNA and disease phenotypes through experiments. High throughput refers to the use of a new generation of high-throughput sequencing technology to sequence lncRNA, and the combination of bioinformatics prediction tools to analyze lncRNA functions.
References
1. Xie, C., Yuan, J., Li, H., Li, M., Zhao, G., Bu, D., Zhu, W., Wu, W., Chen, R. and Zhao, Y. (2014) NONCODEv4: exploring the world of long non-coding RNA genes. Nucleic acids research, 42, D98-D103.
2. Safran, M., Dalah, I., Alexander, J., Rosen, N., Iny Stein, T., Shmoish, M., Nativ, N., Bahir, I., Doniger, T. and Krug, H. (2010) GeneCards Version 3: the human gene integrator. Database, 2010.
3. Yates, A.D., Achuthan, P., Akanni, W., Allen, J., Allen, J., Alvarez-Jarreta, J., Amode, M.R., Armean, I.M., Azov, A.G. and Bennett, R. (2020) Ensembl 2020. Nucleic acids research, 48, D682-D688.
4. Tweedie, S., Braschi, B., Gray, K., Jones, T.E., Seal, R.L., Yates, B. and Bruford, E.A. (2021) Genenames. org: the HGNC and VGNC resources in 2021. Nucleic acids research, 49, D939-D946.
5. Benson, D.A., Karsch-Mizrachi, I., Lipman, D.J., Ostell, J., Rapp, B.A. and Wheeler, D.L. (2000) GenBank. Nucleic acids research, 28, 15-18.
6. Wheeler, D.L., Barrett, T., Benson, D.A., Bryant, S.H., Canese, K., Chetvernin, V., Church, D.M., DiCuccio, M., Edgar, R. and Federhen, S. (2007) Database resources of the national center for biotechnology information. Nucleic acids research, 36, D13-D21.
7. Huang, H.-Y., Lin, Y.-C.-D., Cui, S., Huang, Y., Tang, Y., Xu, J., Bao, J., Li, Y., Wen, J. and Zuo, H. (2022) miRTarBase update 2022: an informative resource for experimentally validated miRNA–target interactions. Nucleic acids research, 50, D222-D230.
8. Ning, S., Zhang, J., Wang, P., Zhi, H., Wang, J., Liu, Y., Gao, Y., Guo, M., Yue, M. and Wang, L. (2016) Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic acids research, 44, D980-D985.
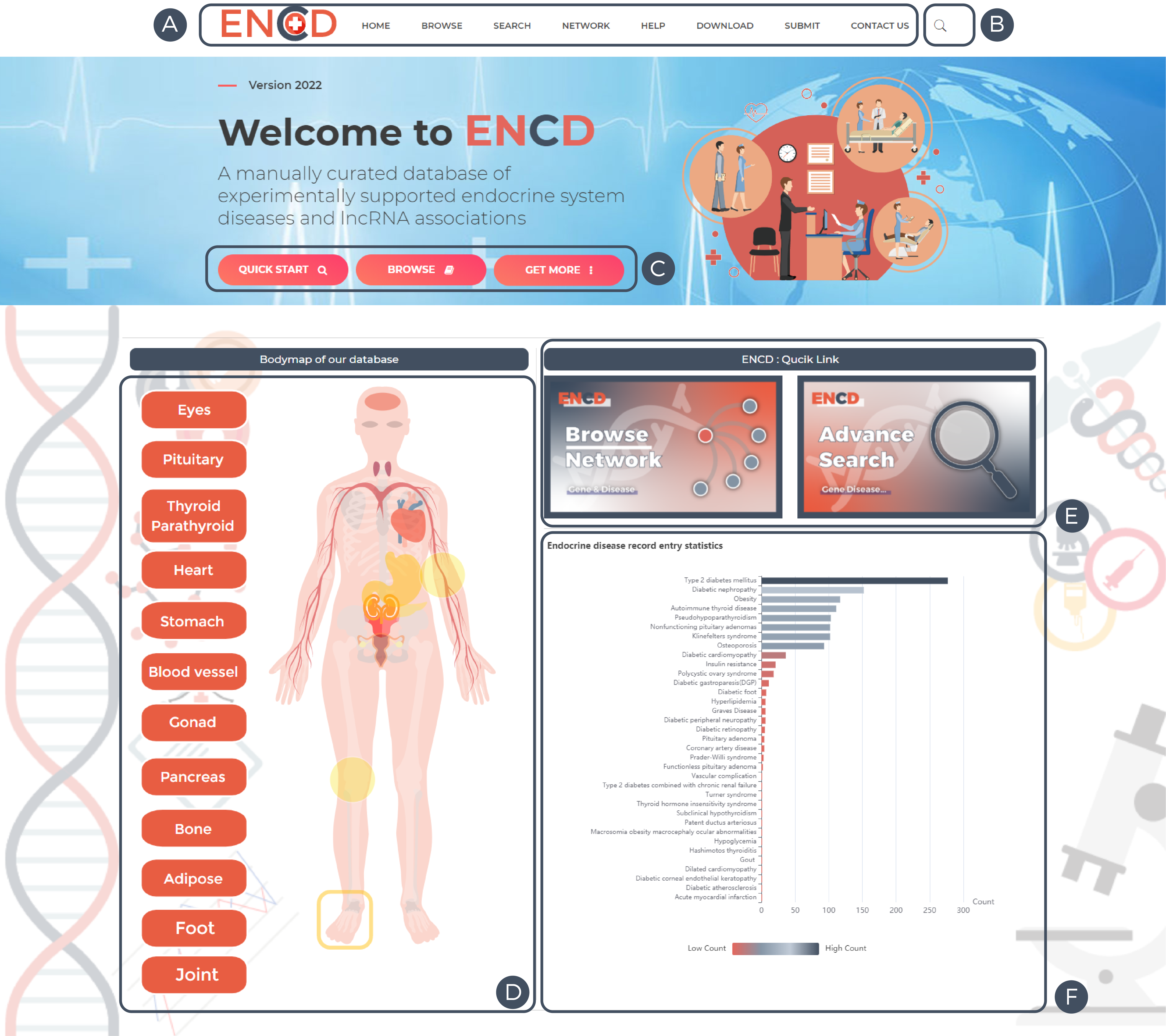
A. ENCD database navigation bar: click on different links to quickly access other ENCD pages.
B. ENCD database quick query function: click this button to bring up the quick query interface.
C. Click the quick start/browse button to enter the search/browse interface.
D. ENCD bodymap: The onset site or the corresponding name queries all diseases contained in that site.
E. ENCD Quick links: Click to visit other ENCD pages.
F. ENCD bar chart: shows the number of relevant lncRNAs for different diseases in this database.
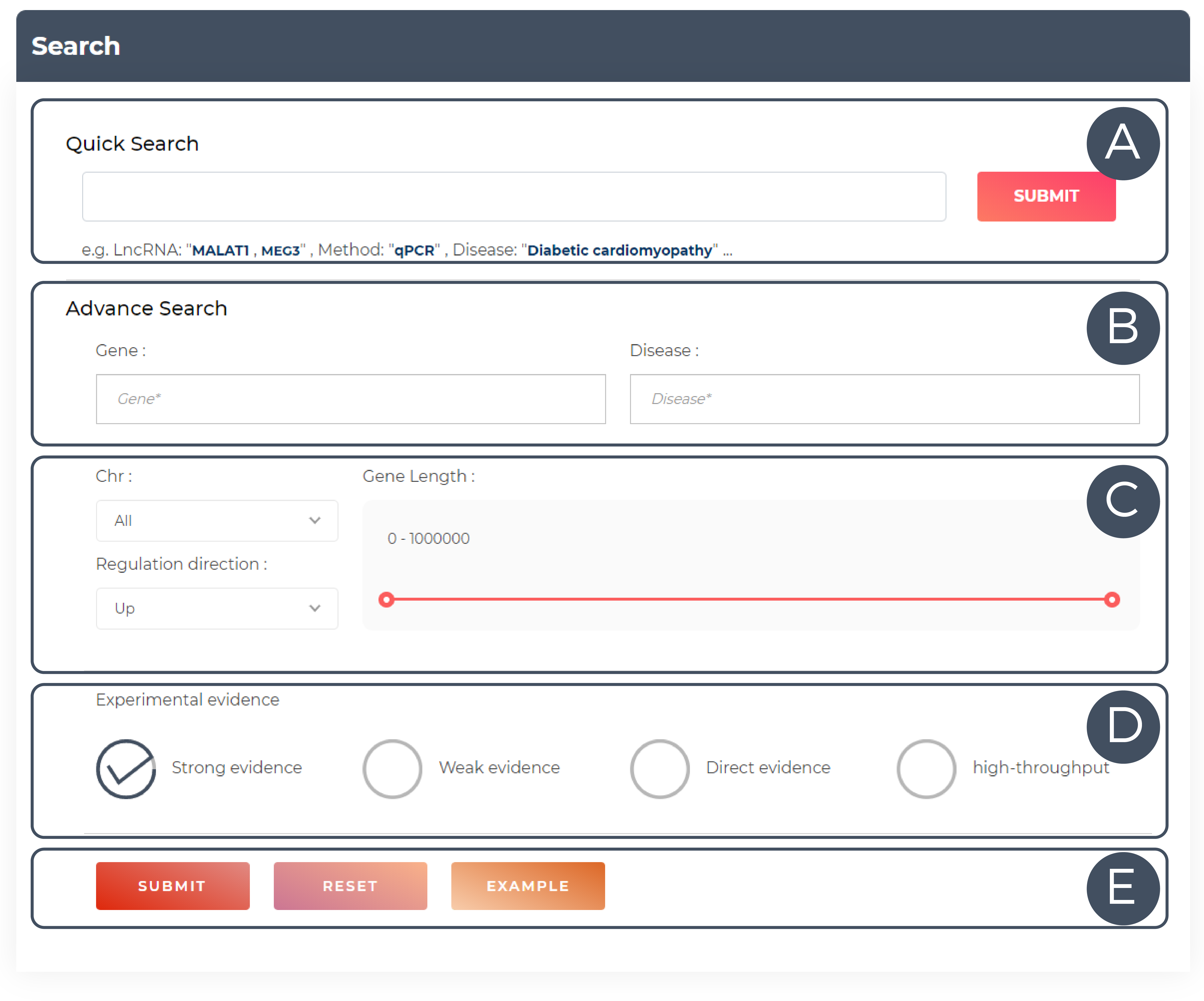
A. Use the Quick Search function to search. Click on the keyword below the input field for quick input (the data field needs to be filled in with a keyword to be able to query the data); and submit by clicking on the SUBMIT button.
B. Advanced search for diseases and genetic data fields (optional).
C. Other information in the conditional screening box, which includes chromosome location, the regulatory role of genes in disease, gene length, etc.
D. The data is confirmed by what type of experiment.
E. Submit/Reset/Example button.
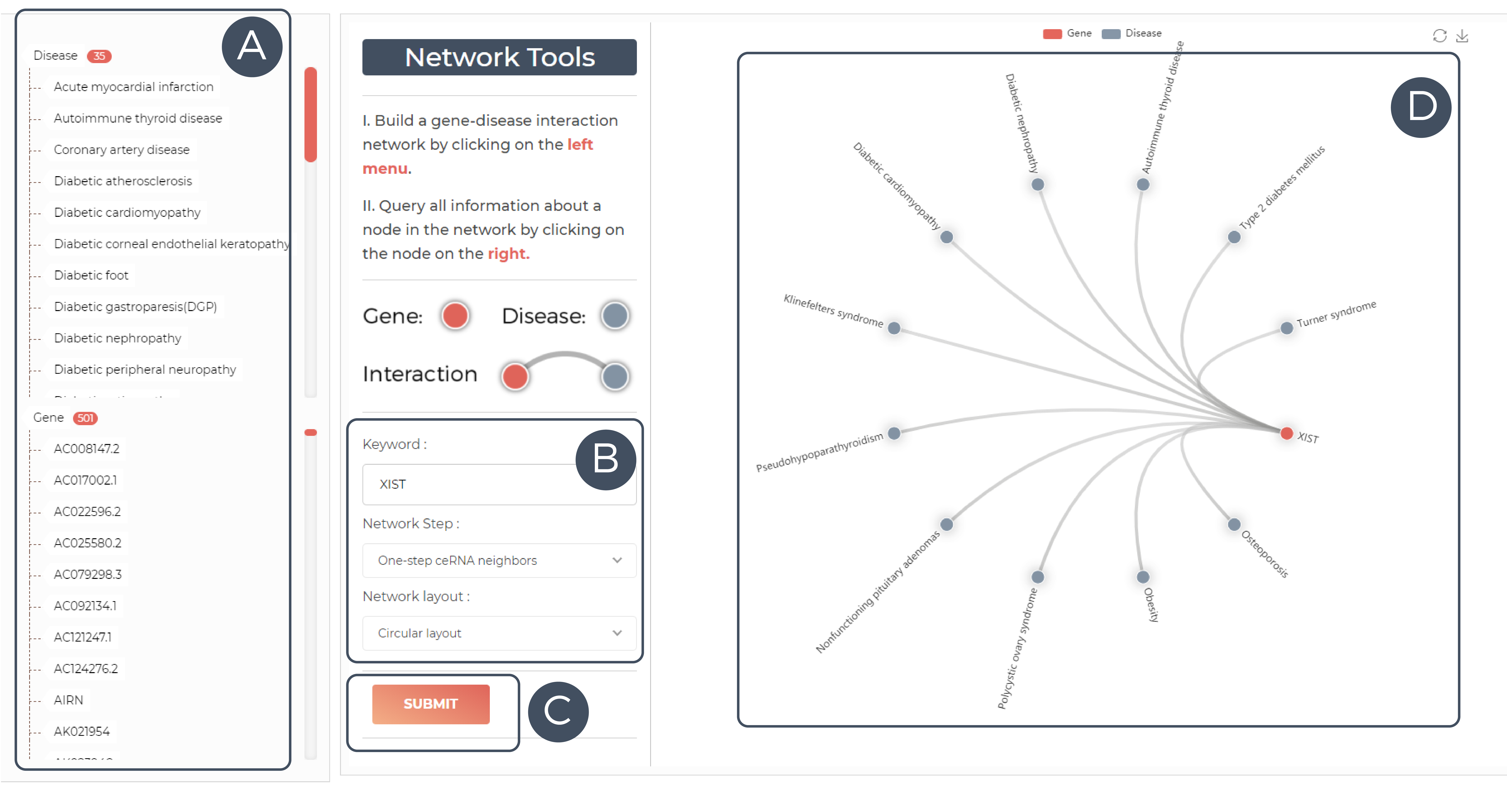
A. All genes & diseases entries in our database.
B,C. Network builder: Change the layout as well as the network step and click the submit button to update the network.
D. Interworking network diagram: Click on one of the nodes to view details.
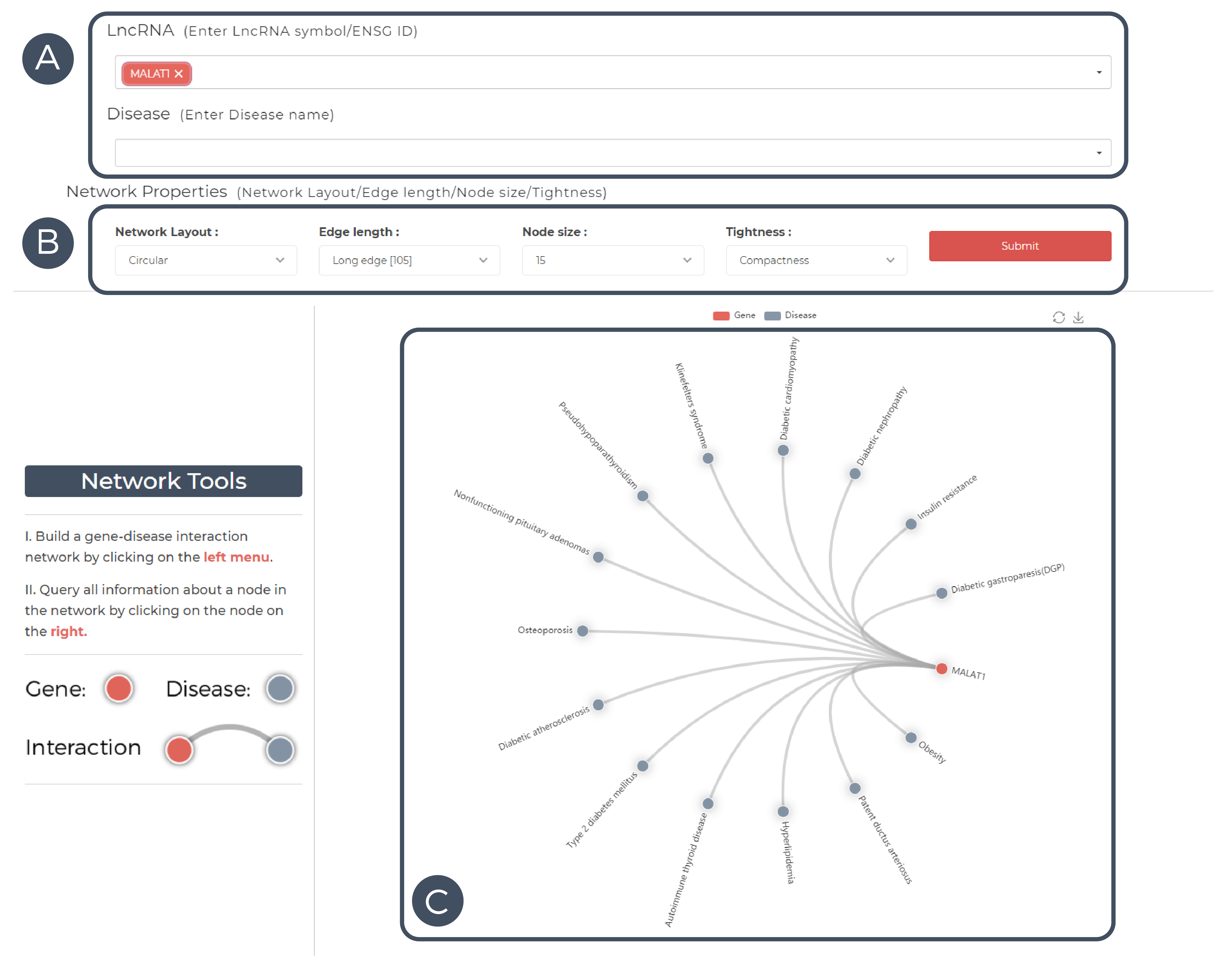
A. Enter LncRNA/disease in the input box.
B. Adjust network properties.
C. Network Diagram Results.
A. Click on the file name to get the corresponding file .

A. Upload your files and click submit button.
B. Describe your detailed question and submit it to us.
A. Our Contact Information.
B. Please provide your questions or suggestions for improvement.